At the 2025 Bio-IT World Conference & Expo in Boston, Dr. Sehyun Oh, Assistant Professor at the City University of New York (CUNY) School of Public Health and Health Policy, delivered a presentation that resonated with a pressing challenge in biomedical R&D:
How do we unlock the full potential of public omics data for AI, machine learning, and translational research?
The answer lies in metadata harmonization, a cornerstone of the FAIR data movement (Findable, Accessible, Interoperable, and Reusable). But FAIRification isn’t a simple checkbox. It requires smart infrastructure, automation, and collaboration across sectors.
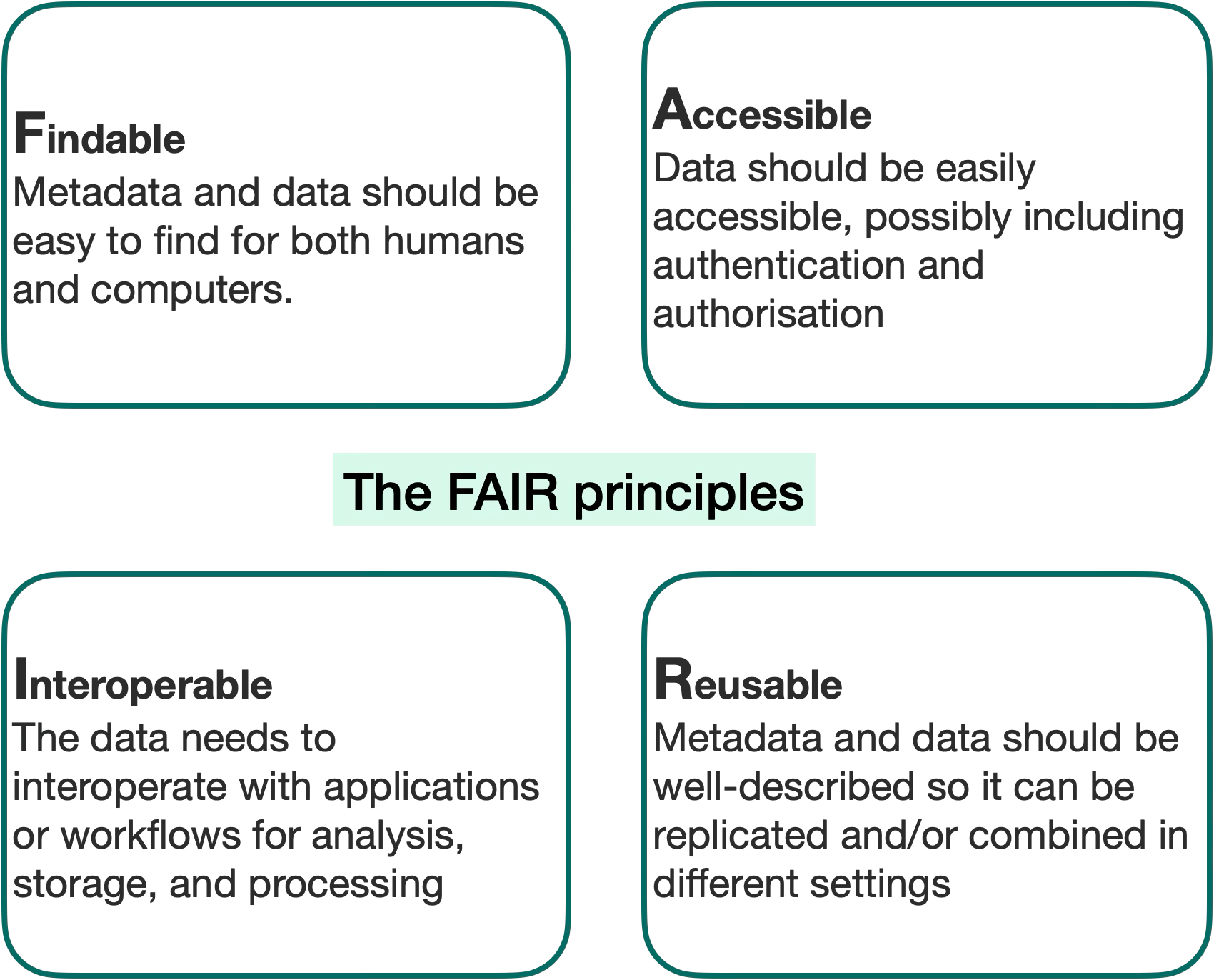
[Figure 1] A diagram showing the definition of FAIR
The Problem: Vast Repositories, Limited Usability
Despite large-scale national efforts to collect and share biological datasets, researchers and companies alike struggle to reuse this data effectively. One of the main reasons is the metadata fragmentation and inconsistency. Without harmonized metadata, even the best high-quality datasets remain hard to discover, integrate, or analyze, especially in AI/ML pipelines.
The challenge is not abstract, it is measurable. A comparative analysis of original versus manually curated metadata reveals just how noisy and inconsistent public omics data can be.
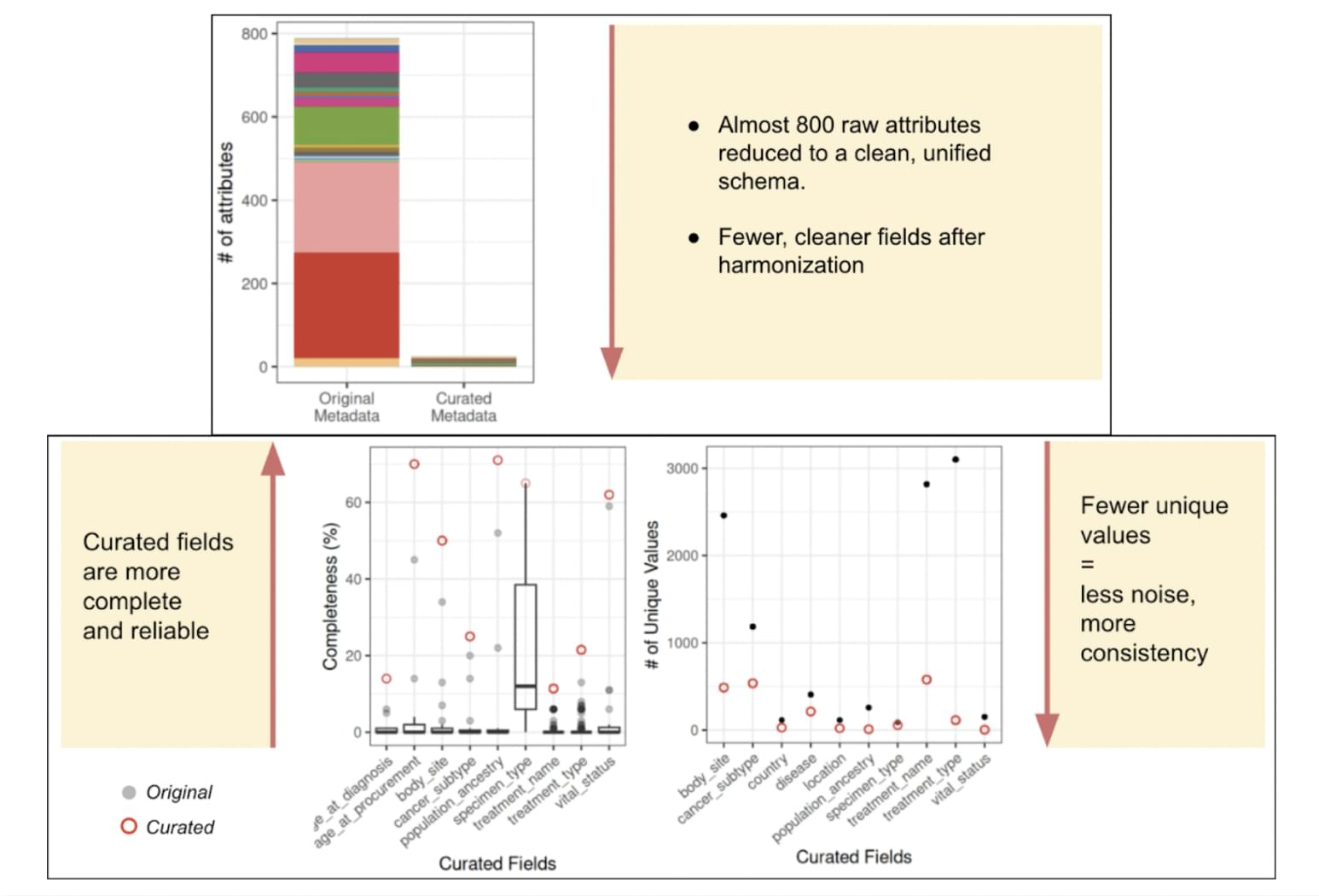
[Figure 2] Impact of metadata harmonization: (Top) Nearly 800 heterogeneous raw attributes from publicly available datasets are reduced to a small, clean set of curated fields. (Bottom left) Curated metadata shows significantly higher completeness. (Bottom right) Curated values are more consistent, with fewer unique entries, enabling better downstream integration.
These results were achieved using manual metadata curation, which is highly effective but also labor-intensive and unsustainable at scale. With data volume constantly growing, the question becomes: How can we automate this process and still ensure quality?
The Solution: OmicsMLRepoR and a FAIR-first Approach
Dr. Oh leads the project OmicsMLRepoR1, focusing on public metagenomics and cancer genomics datasets. The goal of this project is to improve metadata quality and ontology alignment, creating a discoverable and interoperable layer on top of existing repositories.
OmicsMLRepoR1 is part of the Bioconductor2 project, one of the most widely used open-source ecosystems in bioinformatics, making harmonized metadata accessible to researchers via R-based tools.
This enables researchers to ask powerful questions like:
“Find all studies involving colorectal cancer, microbiome profiling, and patient survival outcomes.”
To support this functionality, OmicsMLRepoR combines:
- Consolidated metadata schemas across repositories
- Controlled vocabularies and domain ontologies
- Integration with R/Bioconductor packages
Automating Harmonization: A Multi-Layered Approach
To scale beyond manual curation, OmicsMLRepoR2 uses a multi-stage semantic matching pipeline, processing from simple exact matches to advanced large language model (LLM) techniques for context-aware harmonization.
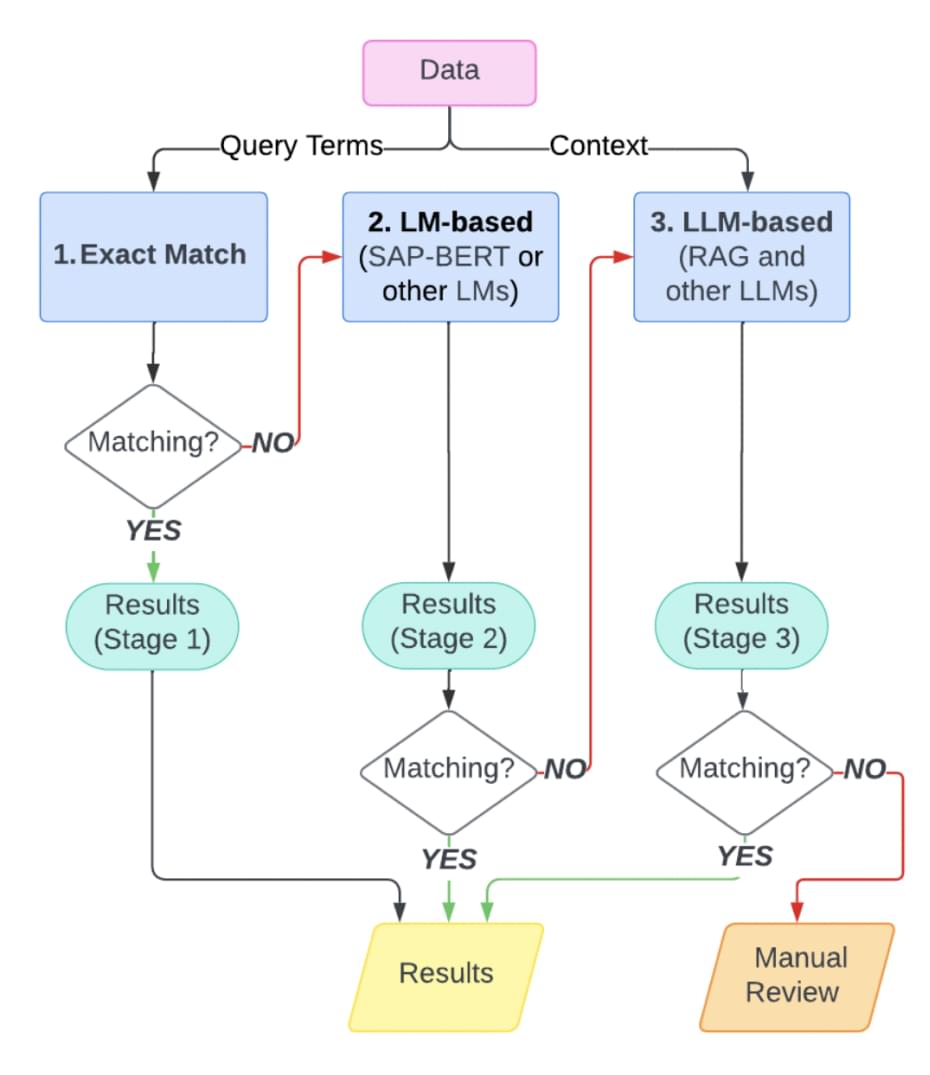
[Figure 3] Multi-layered metadata harmonization pipeline used by OmicsMLRepoR combining exact matching, language models (e.g., SAP-BERT), and large language models (e.g., RAG) for semantic alignment, with manual review as the final quality control3
This layered system ensures that diverse terms across studies, such as labels, specimen types, or treatment data, can be harmonized in a consistent, scalable, and FAIR-compliant way.
Our Collaboration: Genestack’s Role in Scalable FAIRification
Genestack partnered with Dr. Oh to operationalize this approach through our Open Data Manager (ODM) platform. Together, we:
- Built ontology-driven templates to standardize heterogeneous metadata
- Enabled automated ingestion and harmonization of public study descriptors
- Developed scalable curation workflows to ensure consistency and queryability
- Explored integration with Bioconductor, enabling researchers to access harmonized metadata programmatically
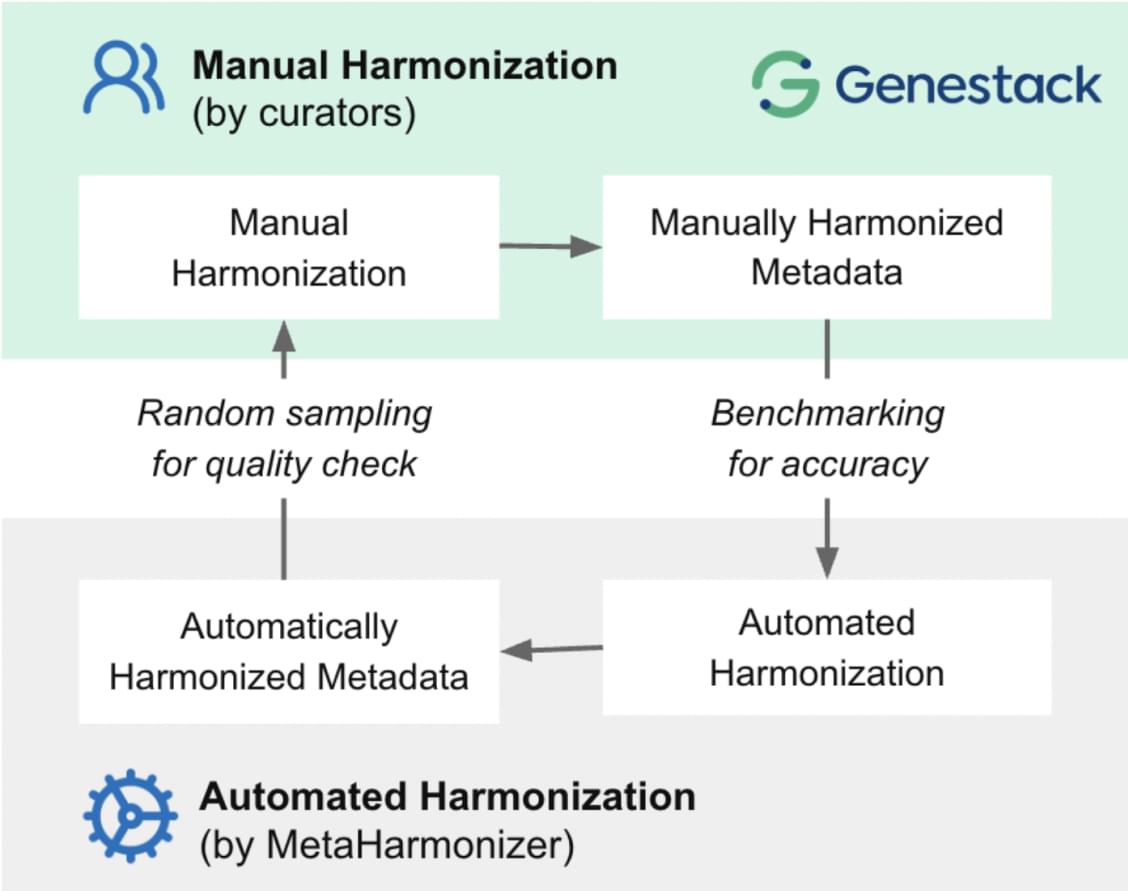
[Figure 4] The collaboration enables harmonization at two levels: (1) consistent quality control of automated processes and (2) repository-specific optimization using a manually curated gold standard
For Genestack, this collaboration represents more than a use case. It is part of our mission to enable FAIRification at scale across public and proprietary datasets.
Why It Matters: Industrial Implications of FAIR Metadata
The Genestack–CUNY collaboration demonstrates that even highly variable public datasets can be transformed into structured, AI/ML-ready assets.
This work is directly applicable to:
- Pharma and biotech teams looking to reuse public datasets alongside internal studies
- Data science teams struggling with metadata chaos
- R&D groups building discovery platforms or federated data infrastructures
By combining academic innovation and industrial-grade data management, we accelerate both open research and commercial applications.
Takeaways from Bio-IT and the Collaboration
In our follow-up call with Dr. Oh after Bio-IT, she emphasized that:
“Without harmonized metadata, it’s nearly impossible to compare datasets, let alone build AI models that generalize.”
Her key takeaways:
- Invest early in metadata modelling and ontology planning
- Encourage academic-industry collaboration to align infrastructure with evolving research needs
- Use platforms like Genestack ODM to reduce manual burden and avoid reinventing curation processes
Looking Ahead: Scaling FAIRification Together
This is just the beginning. Together, we are working toward:
- Seamless Interoperability with Bioconductor and other tools
- Better support for federated data discovery, without duplicating datasets
- Developing reusable curation frameworks to help others harmonize their data more easily
If your organization is facing metadata challenges, whether in internal data lakes or public repositories, we’d love to share what we’ve learned.
Let’s make omics data not just open, but truly usable.

[Figure 5] Sehyun Oh, Professor at CUNY, presenting at BioIT
References
- Oh S, Long K (2025). OmicsMLRepoR: Search harmonized metadata created under the OmicsMLRepo project. https://doi.org/10.18129/B9.bioc.OmicsMLRepoR, R package version 1.2.0, https://bioconductor.org/packages/OmicsMLRepoR.
- Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B., Dettling, M., Dudoit, S., ... & Huber, W. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biology, 5(10), R80. DOI: https://doi.org/10.1186/gb-2004-5-10-r80
- Oh, S. (2025, April 3). Improving FAIRness of Omics Data through Metadata Harmonization [Conference presentation].BioIT World 2025, Boston, MA, United States